A sequel, only better
outbreaker2 is a total re-implementation of a model implemented in outbreaker, for the reconstruction of transmission trees from densely sampled outbreaks (i.e., most cases observed). While the original version was computationally efficient, it basically consisted of a C engine which was optimized for speed, but poorly tested (no unit testing) and very hard to develop upon.
This is unfortunate, as the original model was fairly simple and did not allow addition of new types of data or likelihood components. The original idea behind outbreaker2 was to address these limitations.
What is new?
outbreaker after the yoga revolution
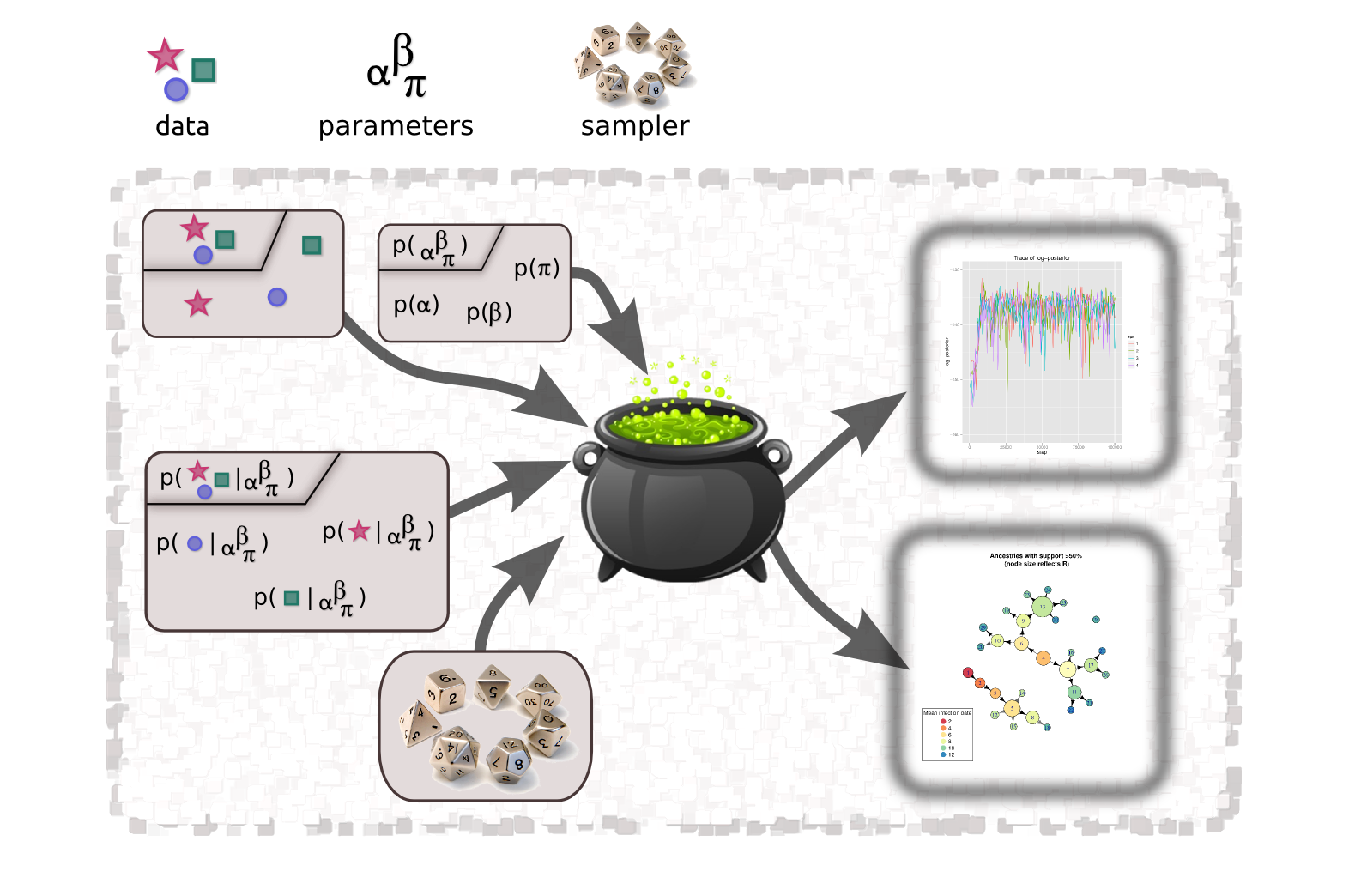
The major novelty of outbreaker2 is flexibility. The design of outbreaker2 relied on breaking down the components making up outbreaker and similar methods, and treating them as separate modules. This includes priors, likekelihood functions, and even movement of parameters and augmented data. All of these modules are treated as separate ingredients of a big methodological recipe, and outbreaker2 provides the cooking pot.
In fact, it also provides the kitchen, as it implements an infrastructure for handling data, defining settings of the method, and customising functions for priors, likelihoods, and random walks in the parameter space. This gives the user more control over the analysis, but will also drastically facilitate the development of new models without risking to break the existing infrastructure. Even if the components of the default outbreaker2 model all runs in C++ (integrated via Rcpp), users can specify new components of the model as R functions, tapping the C++ API where relevant.
Towards more reliability
Another side-benefit of this modularity is making testing simpler. Like most if not all similar methods in the field, outbreaker had no unit testing. Not one. Nothing. outbreaker2 will follow the standards defined by RECON for package development, including complete coverage of the code (in R as well as C++) by unit testing (current coverage is about 70%) and the use of continuous integration tools (currently Travis and Appveyor).
Slower, but better
outbreaker2 is slower than outbreaker by about one order of magnitude, but speed can be deceptive. While the flexibility acquired in outbreaker2 undoubtedly has a cost in terms of speed, outbreaker2 also does considerably more per iteration of the MCMC than its predecessor. For instance, only a fraction of the transmission tree was altered (“moved”) at each MCMC iteration in outbreaker. The same was true of dates of infections, and some other movements. In contrast, outbreaker2 everything that can be moved at each iteration, resulting in faster mixing. It may be 12 times slower, but it mixes approximately 10 times faster, so that the speed difference is not as bad as it seems.
Can I try it?
outbreaker2 has now entered its testing phase. It is fully documented at this website:
http://www.repidemicsconsortium.org/outbreaker2/
The first place to start is the introductory vignette](http://www.repidemicsconsortium.org/outbreaker2/articles/introduction.html).
Feedback is welcome! Feel free to post your comments on the bottom of this page, or on the github issues of the project.