Example to estimate incubation period
Flavio Finger
2019-05-13
estimate_incubation.RmdDescription
This package contains two functions useful to compute the incubation period distribution from outbreak data. The inputs needed for each patient are given as a data.frame or linelist object and must contain:
- the date of onset of symptoms
- a list/vector of possible dates of exposure.
The function empirical_incubation_dist() computes the discrete probability distribution by giving equal weight to each patient. Thus, in the case of N patients, the n possible exposure dates of a given patient get the overall weight 1/(n*N). The function returns a data frame with column incubation_period containing the different incubation periods with a time step of one day and their relative_frequency.
The function fit_gamma_incubation_dist() takes the same inputs, but directly samples from the empirical distribution and fits a discrete gamma distribution to it by the means of fit_disc_gamma.
Example
Load environment:
Make a linelist object containing toy data with several possible exposure dates for each case:
ll <- messy_data() %>% clean_data()
x <- 0:15
y <- distcrete("gamma", 1, shape = 12, rate = 3, w = 0)$d(x)
mkexposures <- function(i) i - sample(x, size = sample.int(5, size = 1), replace = FALSE, prob = y)
exposures <- sapply(ll$date_of_onset, mkexposures)
ll$dates_exposure <- exposures
print(ll)
#> id date_of_onset discharge gender epi_case_definition messy_dates
#> 1 ujfkwq 2018-01-03 2018-01-13 female not_a_case 2001-12-01
#> 2 eyxzua 2018-01-07 2018-01-17 male confirmed <NA>
#> 3 doulii 2018-01-05 2018-01-15 male not_a_case <NA>
#> 4 owvlbu 2018-01-02 2018-01-12 female confirmed <NA>
#> 5 wqbbyq 2018-01-05 2018-01-15 female suspected <NA>
#> 6 mauyjn 2018-01-02 2018-01-12 male not_a_case 2001-12-01
#> 7 wahzsb 2018-01-09 2018-01-19 male probable <NA>
#> 8 wyhlwc 2018-01-02 2018-01-12 female suspected <NA>
#> 9 tbnecx 2018-01-02 2018-01-12 male suspected <NA>
#> 10 brithg 2018-01-08 2018-01-18 male confirmed 2018-10-18
#> 11 kfezyj 2018-01-08 2018-01-18 male suspected <NA>
#> 12 anykpl 2018-01-02 2018-01-12 female probable 1989-12-24
#> 13 ndffve 2018-01-09 2018-01-19 female not_a_case 2001-12-01
#> 14 qykakh 2018-01-09 2018-01-19 male not_a_case <NA>
#> 15 aprglr 2018-01-06 2018-01-16 female not_a_case <NA>
#> 16 hwepnc 2018-01-07 2018-01-17 female probable 2018-10-18
#> 17 nlmqas 2018-01-08 2018-01-18 female probable 2018-10-17
#> 18 wuzznf 2018-01-06 2018-01-16 male suspected <NA>
#> 19 obzshh 2018-01-04 2018-01-14 male suspected 1989-12-24
#> 20 nvlxer 2018-01-04 2018-01-14 female suspected 2018-10-17
#> lat lon dates_exposure
#> 1 12.688208 48.36797 17530, 17529, 17531
#> 2 9.496732 48.10133 17535, 17536
#> 3 13.276658 49.10333 17533, 17532, 17535, 17527, 17534
#> 4 15.443281 50.26166 17529, 17530, 17528, 17531
#> 5 12.186121 47.46422 17534, 17533, 17532, 17531
#> 6 10.887826 48.27850 17528, 17531, 17529, 17530
#> 7 11.755842 47.69728 17539
#> 8 12.484501 50.46869 17528, 17527, 17531
#> 9 11.566569 47.99664 17528, 17529, 17530
#> 10 13.635864 48.10328 17537, 17534, 17535, 17536, 17533
#> 11 15.703823 48.67810 17536
#> 12 12.643401 48.12971 17531, 17530, 17529
#> 13 15.226172 48.88853 17536, 17537, 17538, 17535
#> 14 13.603475 46.06085 17538
#> 15 10.708423 48.01061 17534, 17533, 17531, 17535, 17536
#> 16 14.720354 47.77516 17535, 17534, 17533, 17531, 17536
#> 17 12.902306 49.15254 17536
#> 18 14.379251 46.48828 17535, 17534, 17532, 17530
#> 19 14.677445 47.84708 17532, 17533, 17531, 17530, 17529
#> 20 15.894964 48.53582 17532, 17531, 17530, 17533, 17529Empirical distribution:
incubation_period_dist <- empirical_incubation_dist(ll, date_of_onset, dates_exposure)
print(incubation_period_dist)
#> # A tibble: 10 x 2
#> incubation_period relative_frequency
#> <dbl> <dbl>
#> 1 0 0
#> 2 1 0.0700
#> 3 2 0.231
#> 4 3 0.298
#> 5 4 0.16
#> 6 5 0.152
#> 7 6 0.0567
#> 8 7 0.0225
#> 9 8 0
#> 10 9 0.01
ggplot(incubation_period_dist, aes(incubation_period, relative_frequency)) +
geom_col()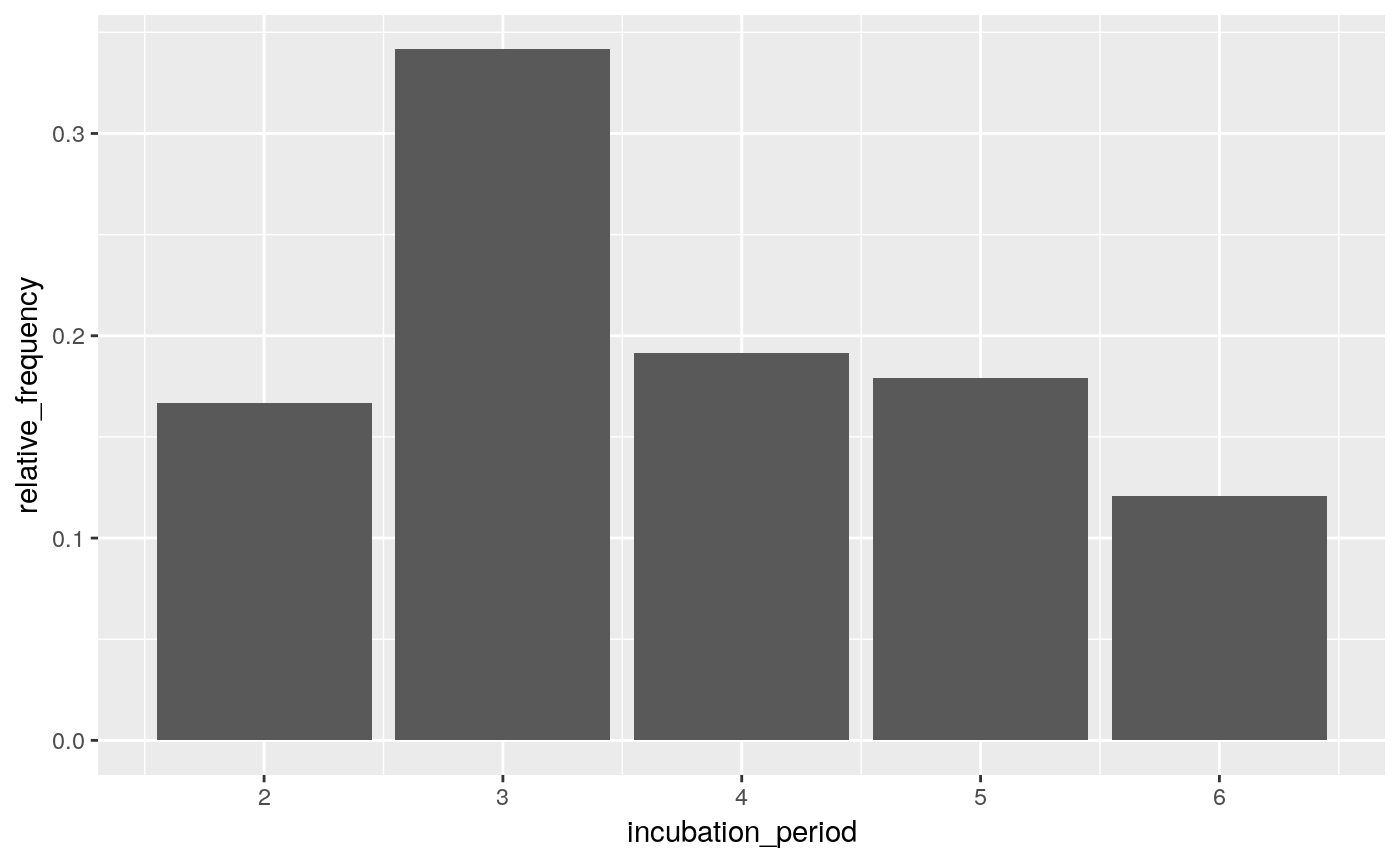
Fit discrete gamma:
fit <- fit_gamma_incubation_dist(ll, date_of_onset, dates_exposure)
print(fit)
#> $mu
#> [1] 3.984821
#>
#> $cv
#> [1] 0.3827553
#>
#> $sd
#> [1] 1.525211
#>
#> $ll
#> [1] -1812.963
#>
#> $converged
#> [1] TRUE
#>
#> $distribution
#> A discrete distribution
#> name: gamma
#> parameters:
#> shape: 6.82586216220056
#> scale: 0.58378274853449
x = c(0:10)
y = fit$distribution$d(x)
ggplot(data.frame(x = x, y = y), aes(x, y)) +
geom_col(data = incubation_period_dist, aes(incubation_period, relative_frequency)) +
geom_point(stat="identity", col = "red", size = 3) +
geom_line(stat="identity", col = "red")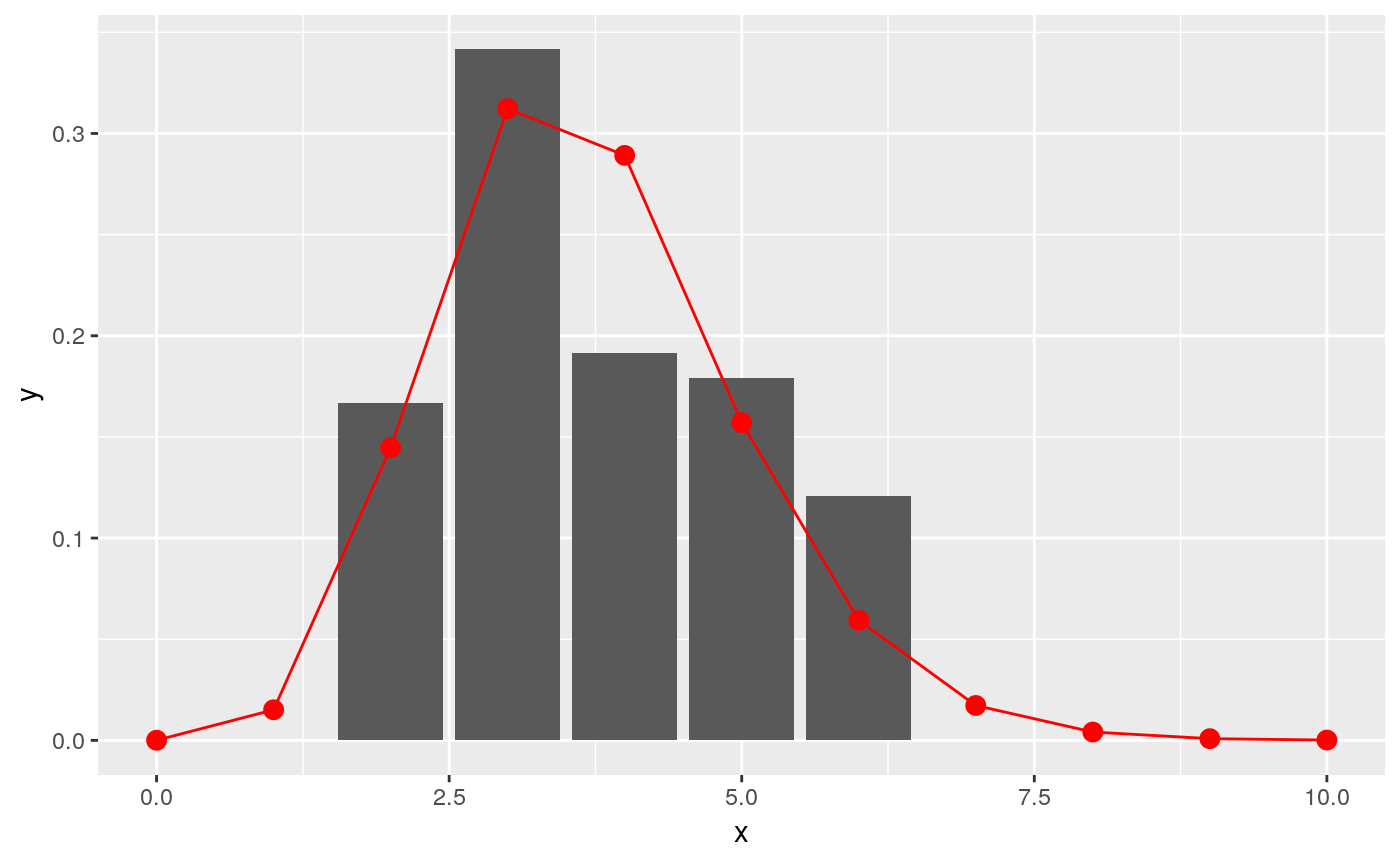
Note that if the possible exposure dates are consecutive for all patients then empirical_incubation_dist() and fit_gamma_incubation_dist() can take date ranges as inputs instead of lists of individual exposure dates (see help for details).